A team of scientists led by researchers from the University of Ghana has developed the Buruli ulcer drug discovery database known as BuDb.
BuDb is the first of its kind and it will aid the search for new treatment options for Buruli ulcer disease. BuDb is published in the Journal of Computational Biophysics and Chemistry and is freely accessible online at http://197.255.126.13:3000/.
The team was led by Dr Samuel Kojo Kwofie of the Department of Biomedical Engineering and the West African Centre for Cell Biology of Infectious Pathogens (WACCBIP), Prof. Michael David Wilson of the Noguchi Memorial Institute for Medical Research (NMIMR), and Dr Lydia Mosi of the Department of Biochemistry, Cell and Molecular Biology and WACCBIP.
Buruli ulcer (BU) is a chronic neglected tropical skin disease caused by Mycobacterium ulcerans. Individuals affected by the disease have skin ulcers. BU is one of the understudied and neglected diseases, and its transmission is yet to be fully elucidated. It is generally acknowledged that BU is associated with water bodies in the environment.
Presently, BU is reported in over 33 countries in Africa, the Americas, Asia and the Western Pacific. Africa has the lion’s share of the world’s BU burden with more than 95% of all cases recorded in the last decade. Côte d’Ivoire, Ghana, Benin, and Cameroon are the four most endemic nations.
Currently, antibiotic therapy is associated with long treatment duration, which is burdened by reported cases of emerging drug resistance. Even though surgery is an option to excise damaged skin and tissue, it can be prohibitively expensive and inaccessible in many countries. Patients oftentimes develop psychological traumas due to the debilitating nature of the scars and joint contractures.
Most essential BU-related data necessary for drug discovery are scattered in published journals and generalized large databases, making searches for standardized and formatted biomedical data necessary to aid drug discovery tedious and time-consuming.
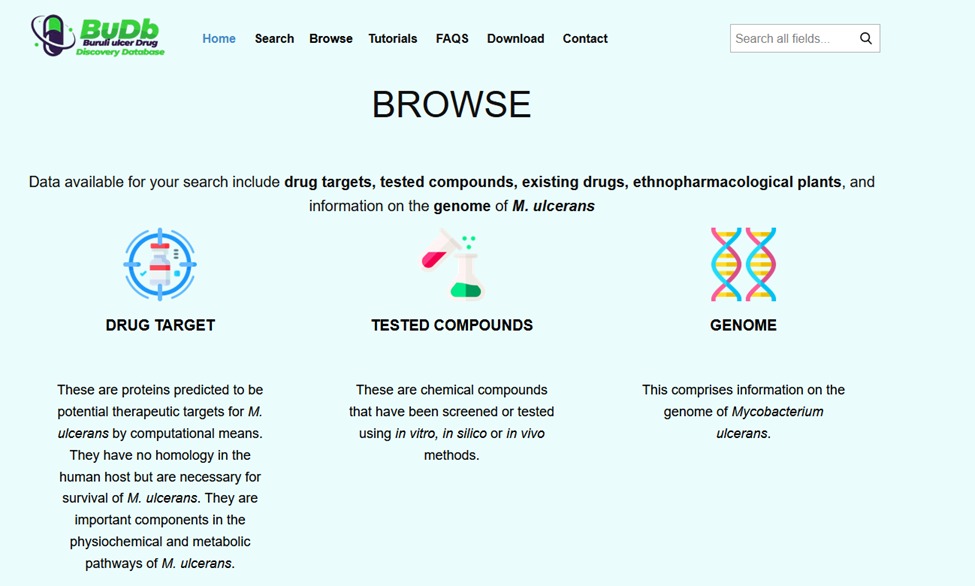
Considering this, a more pragmatic trend has emerged, which is to develop disease-specific databases. BuDb serves as a one-stop knowledge base and a dedicated resource for BU drug discovery. BuDb integrates current drugs under investigation, tested compounds, and traditional treatment options including herbal preparations, drug targets, and information on the genome of M. ulcerans. Understanding the genome allows the identification of new drug targets to which the drugs bind, thereby gaining insights into how the drugs work and designing drugs to overcome the resistance. BuDb integrates links from well-established drug resources and other enriched biological databases allowing cross-referencing of information.
In the era of computer-aided drug discovery, these data are critical elements in the pipeline of novel drug development. The initiative taken by the scientists to provide the first-ever database is a laudable achievement which must be complimented. It is expected that making available the resource to the scientific community will spur the development of new potent Buruli ulcer drugs.
The other team members involved in the biocuration of the data and design of the database are Dr Clement Agoni, Odame Agyapong, Daniel Tweneboah Anyimadu, Solomon Aryee, Blessing Asare, Natalie Kokroko, Jeffrey A. Owusu, Afutu Baron, Edwin Kyei-Baffour, and Kweku Sofo Enninful.





